AI for good: Cleora.AI created by Synerise in Biomedical Sciences.

Artificial Intelligence for drug development in medicine is very hot topic nowadays - which definitely will revolutionize this market segment in the next few years. How can we support this transformation by open sourcing top-notch AI solution which we created in Synerise?
"Drug companies look to AI to end 'hit and miss' research. Artificial intelligence (AI) is set to improve the industry’s success rates and speed up drug discovery, potentially saving it billions of dollars, a recent survey by the analytics firm GlobalData has found. AI topped a list of technologies seen as having the greatest impact on the sector this year. Almost 100 partnerships have been struck between AI specialists and large pharma companies for drug discovery since 2015".
The history of building cleora.ai proves that scientific progress in big data and AI in one field (such as an innovative approach to data processing, analysis and generating insights for commercial companies) can effectively drive other industries, but also affect the progress of civilization in different disciplines such as medicine.
Since the inception of the AI team at Synerise, our ambition has been to quickly and easily process giant heterogeneous data. Existing libraries, such as StarSpace, Node2Vec, DeepWalk, or various graphical convolution networks, did not meet our requirements. Each of them had a drawback, like very slow performance, impractical limitation of the maximum graph size or unsatisfactory quality of the results. We needed a solution that would allow us to quickly and accurately calculate graph embeddings with millions of vertices and billions of edges to represent user behavior. Several months of waiting for the result of the dive was unacceptable for us.The first version of Cleora was created at the beginning of 2019 and was implemented in Scala. It was quickly apparent that the tool successfully replaced all existing embedding libraries.In the next iteration, at the beginning of 2020, in addition to optimizing the algorithm, we decided to get rid of JVM. The entire solution was rewritten from Scala to Rust, thanks to which we have more control over memory and processor consumption, and the performance more than doubled. Its development gave us additional opportunities to create a number of solutions based on it, including to generate recommendations, scoring, segmentation and various predictions.The experience gathered by the entire AI team allowed us to make Cleora what it is today, a universal and reliable "Swiss Army knife" for graph plunging.
Cleora is one of the fastest graph embedding algorithms. It is an essential element for systems operating on data in the form of a network of connected nodes. These are recommendation systems, prediction of connections between users (like / follow) in social media, or systems predicting the biological functions of protein networks, which allows for the creation of new drugs.No wonder then that such algorithms are created by digital giants such as Facebook and Google, creating a number of new solutions each year. However, Cleora has a significant advantage over these algorithms.First, it is much faster. Secondly, it does not require specialized hardware (e.g. GPUs for acceleration of calculations) and, in addition, produces high quality embedding vectors. This means that systems (e.g. referrals) using Cleory may run faster and with greater accuracy.Cleora is capable of processing graphs of hundreds of millions of nodes. In social networks, onenode usually corresponds to a single user, so Cleora can be used to process datasets/graphs on a global scale, at the level of the number of users of the largest social networking sites such as Twitter. The release of the software under an open-source license means that from now on, either a company, an individual or a research institution can use Cleora for any purpose. We recommend Cleora when working with large graphs, especially in conditions of limited computing power. The implementation is available on GitHub. In the scientific sphere, the Synerise team achieved significant success by winning the Rakuten Data Challengecompetition at the SIGIR (Special Interest Group on Information Retrieval) conference. The subject of the competition was creating recommendations in e-commerce, and the organizers included Tracy H. King (Adobe), Shervin Malmasi (Amazon), Dietmar Jannach (University of Klagenfurt), Weihua Luo (Alibaba), Surya Kallumadi (The Home Depot).The Synerise publication on methods for detecting the most important features of products, determining the user's interest, also appeared in the materials of the ICML 2020 conference, and a few months later, at the ICONIP 2020 conference, our article was presented, describing the model selecting similar items of clothing based on a photo gallery from producers and users.
How cleora.ai can help to improve research work on drug discovery?
The progress of research in field of using AI in medicine and drug discovery is amazing. Such amazing scientists like top researchers in graph methods + biomedical: e.g. the SNAP group from Stanford: Jure Leskovec from Stanford University or Marinka Zitnik from Harvard University who are publishing extreme useful materials and educate market about new opportunites (Graph Neural Networks for Drug Development).

Cleora.ai can help achieve and improve daily work of people engaged daily in drug discovery process.
Chemical molecules and cellular structures are graphs, similarly as social networks, road networks and other structures found in nature & human activity:
Thus, processing of molecular/cellular data is inherently a graph processing problem.
Cleora computes graph embeddings – compressed numerical representations of graph nodes, which make it easy and feasible to discover various properties of nodes and whole graphs. By embedding a molecule graph with Cleora, we obtain a versatile representation of the molecule’s properties with can be exploited for multiple purposes.
Cleora embeddings can be used in two ways:
- Directly as source of information - e. g. finding similar nodes/graphs/subgraphs by distance comparison
- As input to more complex method. This way Cleora can enhance system performance by providing informative inputs which facilitate learning.
You can explore great examples here.
Problems solved by Graph Neural Network (GNN) models
Problem1: Find which diseases a chemical compound can treat.
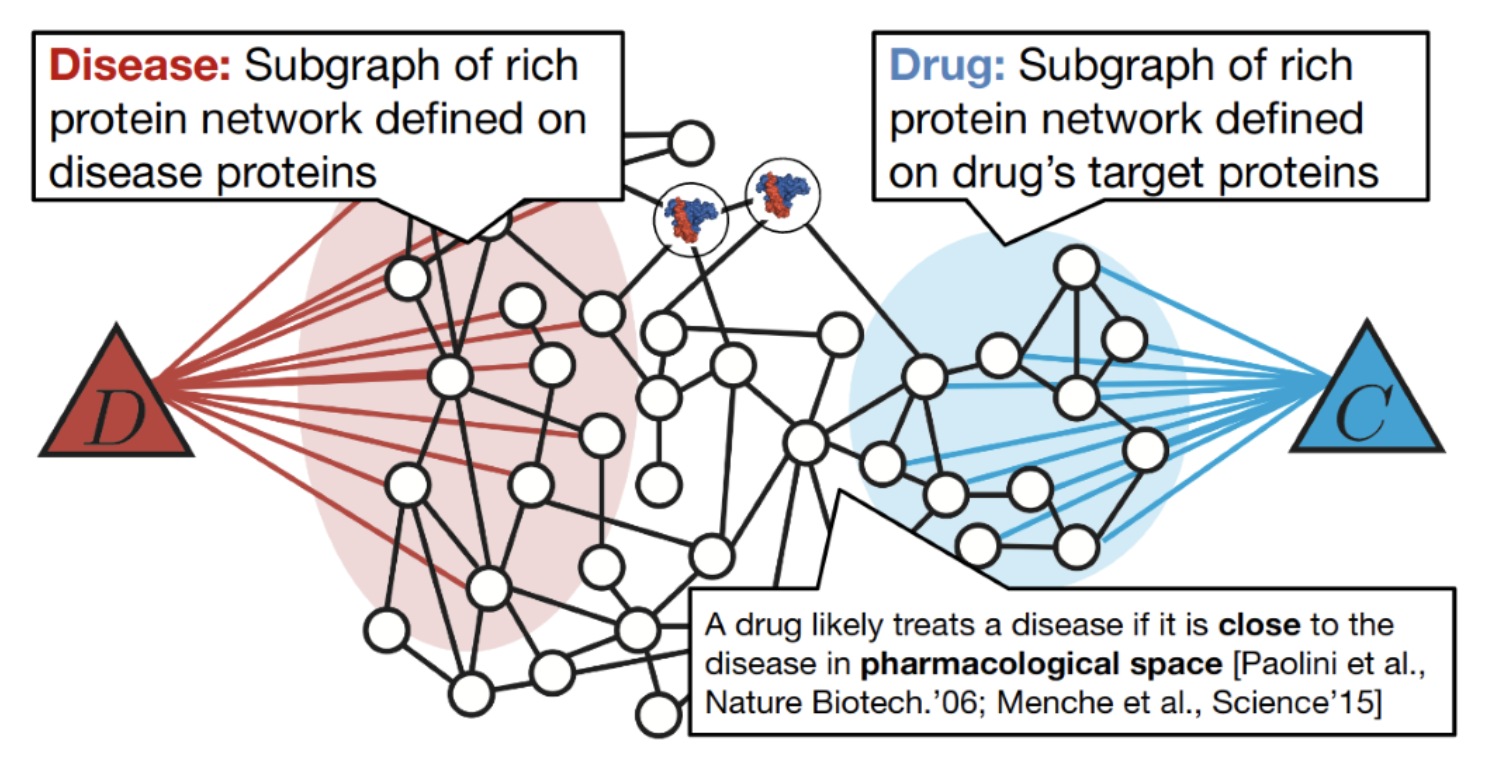
Source: Graph Neural Networks for Drug Development Marinka Zitnik
Solution: subgraph analysis: predict closeness in the graph representation space between drug target protein subgraph and disease protein subgraph (both embedded with Cleora).
Problem 2: Find unexpected drug interactions. Patients take multiple drugs to treat complex or co-existing diseases. 46% of people over 65 years take 5-20 drugs. 15% of the U.S. population affected by unwanted side effects. Annual costs in treating side effects exceed $177 billion in the U.S. alone.
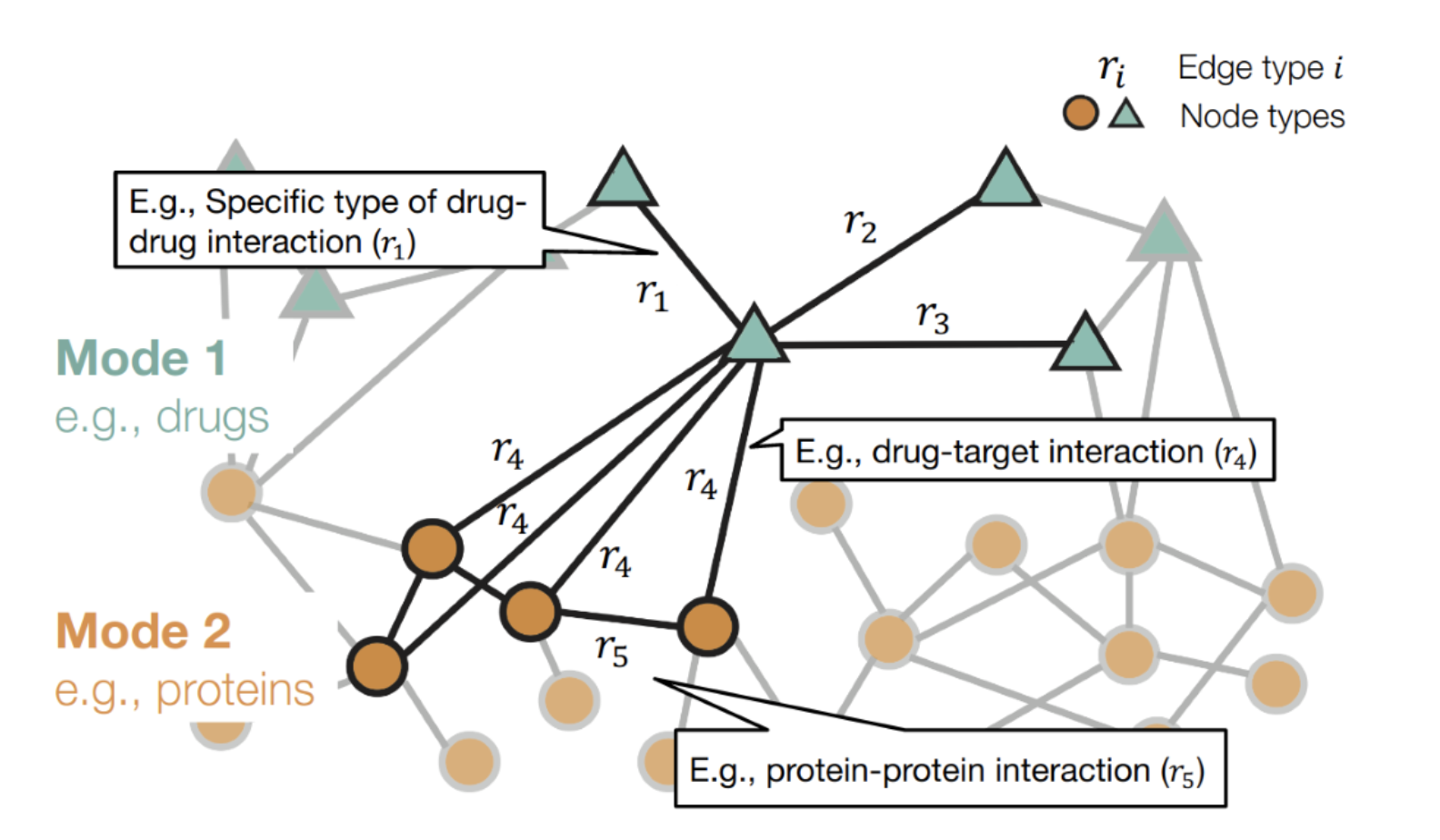
Source:Graph Neural Networks for Drug Development Marinka Zitnik
Solution: Embed drug graphs with Cleora and predict the existence of edges. and predict the existence of edges.
Problem 3: Cell ontologies. Single cell technologies have rapidly generated an unprecedented amount of data that enables us to understand biological systems at single-cell resolution. However, joint analysis of datasets generated by independent labs remains challenging due to a lack of consistent terminology to describe cell types.
https://www.biorxiv.org/content/10.1101/810234v2
Solution: Embed cell ontologies with Cleora, locate similar nodes.
Graph Neural Network Methods - challenges
Problems mentioned above have been previously solved with Graph Neural Network (GNN) models, however, there are some serious problems:
- GNNs are slow and demand significant resources (complexity grows rapidly with increasing graph connectiveness and size)
- An emerging research trend shows that variants of GNNs use superfluous operations which increase complexity but do not bring increased performance
http://proceedings.mlr.press/v97/wu19e.html
https://openreview.net/forum?id=S1ldO2EFPr
https://arxiv.org/abs/1905.04579
The advantages of Cleora
Cleora solves these problems accordingly:
- It only uses the key operation of GNN neighborhood smoothing which is key to performance
- As a result, it is significantly simpler and faster than GNNs
- High scalability and speed allows Cleora to embed graphs with millions of nodes and billions of edges. This is a key feature in biomedical sciences where often all possible patterns of connectivity must be explored, which results in millions of possible configurations.
- It is versatile – it has been applied in winning solutions to competitions from various areas (route prediction, item recommendation, multimodal retrieval)
Barbara Rychalska, Jarosław Królewski
Cleora: A Simple, Strong and Scalable Graph Embedding Scheme -Paper
The area of graph embeddings is currently dominated by contrastive learning methods, which demand formulation of an explicit objective function and sampling of positive and negative examples. This creates a conceptual and computational overhead. Simple, classic unsupervised approaches like Multidimensional Scaling (MSD) or the Laplacian eigenmap skip the necessity of tedious objective optimization, directly exploiting data geometry. Unfortunately, their reliance on very costly operations such as matrix eigendecomposition make them unable to scale to large graphs that are common in today's digital world. In this paper we present Cleora: an algorithm which gets the best of two worlds, being both unsupervised and highly scalable. We show that high quality embeddings can be produced without the popular step-wise learning framework with example sampling. An intuitive learning objective of our algorithm is that a node should be similar to its neighbors, without explicitly pushing disconnected nodes apart. The objective is achieved by iterative weighted averaging of node neigbors' embeddings, followed by normalization across dimensions. Thanks to the averaging operation the algorithm makes rapid strides across the embedding space and usually reaches optimal embeddings in just a few iterations. Cleora runs faster than other state-of-the-art CPU algorithms and produces embeddings of competitive quality as measured on downstream tasks: link prediction and node classification. We show that Cleora learns a data abstraction that is similar to contrastive methods, yet at much lower computational cost. We open-source Cleora under the MIT license allowing commercial use under this https URL.
